项目说明:
根据用户诊断信息数据判断泌尿系统的疾病–肾炎症还是膀胱炎症? 这是一个典型的分类问题
数据集:
http://archive.ics.uci.edu/ml/machine-learning-databases/acute/
a1 病人的体温 { 35C-42C }
a2 有恶心反胃症状 { yes, no }
a3 是否腰疼 { yes, no }
a4 排尿情况是否顺畅 { yes, no }
a5 排尿疼痛 { yes, no }
a6 尿道出口灼烧感,痒或肿胀 { yes, no }
d1 疾病: 肾炎 { yes, no }
d2 疾病: 膀胱炎 { yes, no }
数据样例:
35,5 no yes no no no no no
35,9 no no yes yes yes yes no
35,9 no yes no no no no no
36,0 no no yes yes yes yes no
36,0 no yes no no no no no
36,0 no yes no no no no no
36,2 no no yes yes yes yes no
数据特征分布分析:
df = pd.read_csv(inPath,sep =",",low_memory=False)
df.columns = ['Temp','what','nausea','Lumbar_pain','Urine_pushing','Micturition_pains','Burning_urethra','decision_urinary','decision_renal_pelvis']
print(df.describe())
print(df.describe(include=['O']))
print(df.info())数值型特征分布:
目前都无缺失值,what这个特征类型没有说明,暂且叫what…
Temp what
count 119.000000 119.000000
mean 38.352941 3.983193
std 1.862149 3.135319
min 35.000000 0.000000
25% 37.000000 1.000000
50% 38.000000 4.000000
75% 40.000000 7.000000
max 41.000000 9.000000非数值型特征分布:
nausea ... decision_renal_pelvis
count 119 ... 119
unique 2 ... 2
top no ... no
freq 90 ... 69特征类型分布:
和上面数值型,非数值型分析的情况一致,没有异常类型
Data columns (total 9 columns):
Temp 119 non-null int64
what 119 non-null int64
nausea 119 non-null object
Lumbar_pain 119 non-null object
Urine_pushing 119 non-null object
Micturition_pains 119 non-null object
Burning_urethra 119 non-null object
decision_urinary 119 non-null object
decision_renal_pelvis 119 non-null object空值判断:
total = df.isnull().sum()#.sort_values(ascending=False)
percent = (df.isnull().sum()/df.isnull().count())#.sort_values(ascending=False)x`
missing_data = pd.concat([total, percent], axis=1, keys=['Total', 'Percent'])
print(missing_data)结果显示离线数据暂时都无空值
Total Percent
Temp 0 0.0
what 0 0.0
nausea 0 0.0
Lumbar_pain 0 0.0
Urine_pushing 0 0.0
Micturition_pains 0 0.0
Burning_urethra 0 0.0
decision_urinary 0 0.0
decision_renal_pelvis 0 0.0特征相关性分析:
数值型特征与Label相关性:
所有特征和Label–decision_urinary ,decision_renal_pelvis 做相关性分析
temp VS decision_urinary,temp VS decision_renal_pelvis
print(df[['decision_urinary', 'Temp']].groupby(['decision_urinary'], as_index=False).mean().sort_values(by='Temp', ascending=False))
print(df[['decision_renal_pelvis', 'Temp']].groupby(['decision_renal_pelvis'], as_index=False).mean().sort_values(by='Temp', ascending=False))
温度在37度左右一般是decision_urinary,温度在39+左右一般是decision_renal_pelvis
decision_urinary Temp
0 no 38.833333
1 yes 37.864407
decision_renal_pelvis Temp
1 yes 39.960000
0 no 37.188406其他特征分析类似,what为4.2左右时一般是decision_urinary,what为3.4左右时一般是decision_renal_pelvis
decision_urinary what
1 yes 4.237288
0 no 3.733333
decision_renal_pelvis what
0 no 4.376812
1 yes 3.440000非数值型特征与Label的相关性:
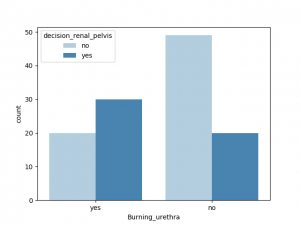
sns.countplot(x = "nausea", hue ="decision_urinary",data = df, palette = "Blues")
plt.show( )
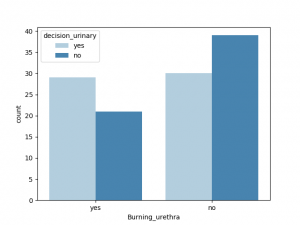
与decision_urinary 相关性最大的非数值是Urine_pushing, 其他特征也有一定相关性,Burning_urethra 与之相关性最小
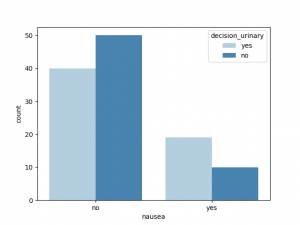
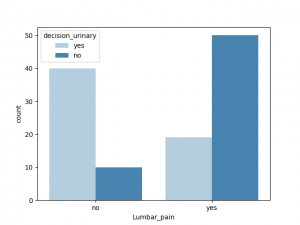
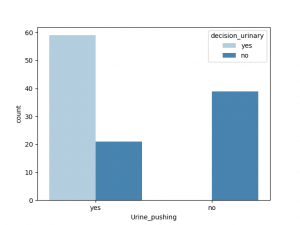
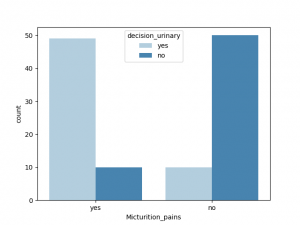
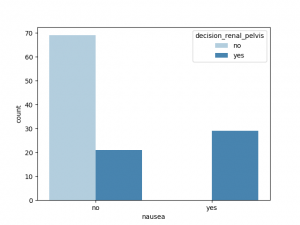
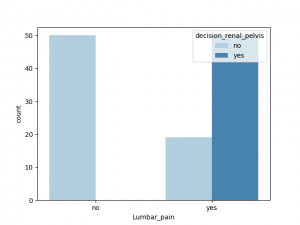
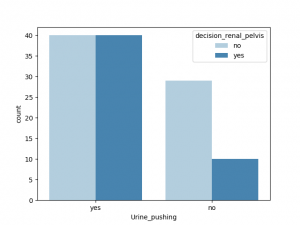
与decision_renal_pelvis 相关性最大的非数值特征是nausea,Lumbar_pain, 其他特征也有一定相关性,Urine_pushing,Micturition_pain 与之相关性较小

特征处理:
根据上述离线数据分析,数据特征处理主要涉及:
a. 非空数据判断 b. 非数值特征数值化--主要是二进制特征数据,比较简单 c. label值处理: 把decision_urinary,decision_renal_pelvis 合并成一个多元变量表示,0表示decision_urinary,1表示decision_renal_pelvis, 2表示这两种病都不是,3表示2中疾病都有
我们假设数据是实时传输过来的,使用Flink 做实时特征处理:
1) 首先定义数据结构类,用于定义数据的导入结构和简单的异常判断:
public class DataDiagnosis implements Serializable {
public int Temp;
public int what;
public String nausea;
public String Lumbar_pain;
public String Urine_pushing;
public String Micturition_pains;
public String Burning_urethra;
public String decision_urinary ;
public String decision_renal_pelvis;
public DateTime eventTime;
public DataDiagnosis() {
this.eventTime = new DateTime();
}
public DataDiagnosis(int Temp, int what, String nausea, String Lumbar_pain,
String Urine_pushing, String Micturition_pains, String Burning_urethra, String decision_urinary,
String decision_renal_pelvis) {
this.eventTime = new DateTime();
this.Temp = Temp;
this.what = what;
this.nausea = nausea;
this.Lumbar_pain = Lumbar_pain;
this.Urine_pushing = Urine_pushing;
this.Micturition_pains = Micturition_pains;
this.Burning_urethra = Burning_urethra;
this.decision_urinary = decision_urinary;
this.decision_renal_pelvis = decision_renal_pelvis;
}
public String toString() {
StringBuilder sb = new StringBuilder();
sb.append(Temp).append(",");
sb.append(what).append(",");
sb.append(nausea).append(",");
sb.append(Lumbar_pain).append(",");
sb.append(Urine_pushing).append(",");
sb.append(Micturition_pains).append(",");
sb.append(Burning_urethra).append(",");
sb.append(decision_urinary).append(",");
sb.append(decision_renal_pelvis);
return sb.toString();
}
public static DataDiagnosis instanceFromString(String line) {
String[] tokens = line.split(",");
if (tokens.length != 9) {
System.out.println("#############Invalid record: " + line+"\n");
return null;
//throw new RuntimeException("Invalid record: " + line);
}
DataDiagnosis diag = new DataDiagnosis();
try {
diag.Temp = tokens[0].length() > 0 ? Integer.parseInt(tokens[0]):null;
diag.what = tokens[1].length() > 0 ? Integer.parseInt(tokens[1]):null;
diag.nausea = tokens[2].length() > 0 ? tokens[2] : null;
diag.Lumbar_pain = tokens[3].length() > 0 ? tokens[3] : null;
diag.Urine_pushing = tokens[4].length() > 0 ? tokens[4] : null;
diag.Micturition_pains = tokens[5].length() > 0 ? tokens[5] : null;
diag.Burning_urethra =tokens[6].length() > 0 ? tokens[6] : null;
diag.decision_urinary = tokens[7].length() > 0 ? tokens[7] : null;
diag.decision_renal_pelvis = tokens[8].length() > 0 ? tokens[8] : null;
} catch (NumberFormatException nfe) {
throw new RuntimeException("Invalid record: " + line, nfe);
}
return diag;
}
public long getEventTime() {
return this.eventTime.getMillis();
}
}流式文件source定义
public class DiagSource implements SourceFunction<DataDiagnosis> {
private final String dataFilePath;
private final int servingSpeed;
private transient BufferedReader reader;
private transient InputStream FStream;
public DiagSource(String dataFilePath) {
this(dataFilePath, 1);
}
public DiagSource(String dataFilePath, int servingSpeedFactor) {
this.dataFilePath = dataFilePath;
this.servingSpeed = servingSpeedFactor;
}
@Override
public void run(SourceContext<DataDiagnosis> sourceContext) throws Exception {
FStream = new FileInputStream(dataFilePath);
reader = new BufferedReader(new InputStreamReader(FStream, "UTF-8"));
String line;
long time;
while (reader.ready() && (line = reader.readLine()) != null) {
DataDiagnosis diag = DataDiagnosis.instanceFromString(line);
if (diag == null){
continue;
}
time = getEventTime(diag);
sourceContext.collectWithTimestamp(diag, time);
sourceContext.emitWatermark(new Watermark(time - 1));
}
this.reader.close();
this.reader = null;
this.FStream.close();
this.FStream = null;
}
public long getEventTime(DataDiagnosis diag) {
return diag.getEventTime();
}
@Override
public void cancel() {
try {
if (this.reader != null) {
this.reader.close();
}
if (this.FStream != null) {
this.FStream.close();
}
} catch (IOException ioe) {
throw new RuntimeException("Could not cancel SourceFunction", ioe);
} finally {
this.reader = null;
this.FStream = null;
}
}
}flink Operation chaim 定义
StreamExecutionEnvironment env = StreamExecutionEnvironment.getExecutionEnvironment();
// operate in Event-time
env.setStreamTimeCharacteristic(TimeCharacteristic.EventTime);
// start the data generator
DataStream<DataDiagnosis> DsDiag = env.addSource(
new DiagSource(input, servingSpeedFactor));
DataStream<DataDiagnosis> modDataStr = DsDiag
.filter(new NoneFilter());
DataStream<String> dataStrForLR = modDataStr.flatMap(new diagFlatMapForRF());
dataStrForLR.print();
dataStrForLR.writeAsText("./diagDataForRF");
env.execute("Taxi Ride Prediction"); 自定义flatmap函数用于生成适合Spark ML RF函数支持的文件格式, 同时把2个label转换成一个多元变量表示:
//使用Spark ML RF, 数据格式保存为csv常见格式
// 2个label 转换成一个多元变量表示
public static class diagFlatMapForRF implements FlatMapFunction<DataDiagnosis, String> {
@Override
public void flatMap(DataDiagnosis InputDiag, Collector<String> collector) throws Exception {
DataDiagnosis diag = InputDiag;
StringBuilder sb = new StringBuilder();
sb.append(diag.Temp).append(",");
sb.append(diag.what).append(",");
sb.append(quantize(diag.nausea)).append(",");
sb.append(quantize(diag.Lumbar_pain)).append(",");
sb.append(quantize(diag.Urine_pushing)).append(",");
sb.append(quantize(diag.Micturition_pains)).append(",");
sb.append(quantize(diag.Burning_urethra)).append(",");
sb.append(newLabel(diag.decision_urinary,diag.decision_renal_pelvis));
collector.collect(sb.toString());
}
private String quantize(String val){
if ("yes".equals(val)){
return "1";
}else if("no".equals(val)){
return "0";
}else{
throw new RuntimeException("Invalid val(must either be 'yes' or 'no'): " + val);
}
}
private String newLabel(String decision_urinary,String decision_renal_pelvis){
if ("yes".equals(decision_urinary) && "no".equals(decision_renal_pelvis)){
return "0";
}else if("yes".equals(decision_renal_pelvis) && "no".equals(decision_urinary)){
return "1";
}else if("no".equals(decision_renal_pelvis) && "no".equals(decision_urinary)){
return "2";
}else if("yes".equals(decision_renal_pelvis) && "yes".equals(decision_urinary)){
return "3";
}else{
throw new RuntimeException("Invalid decision_urinary/decision_renal_pelvis(must either be 'yes' or 'no'): decision_urinary=" + decision_urinary+",decision_renal_pelvis="+decision_renal_pelvis);
}
}
}数据文件格式样例:
3> 41,5,1,1,0,1,0,1
6> 37,2,0,1,0,0,0,2
7> 40,2,1,1,0,1,0,1
8> 37,7,0,1,0,0,0,2
6> 37,5,0,1,0,0,0,2
7> 40,6,1,1,1,1,1,4
模型训练–分类:
使用Spark ML 的批处理任务做模型训练—决策树或者随机森林,看个人喜好
object DiagModRF{
//定义文件读出格式类
case class Diag(
Temp: Double, what: Double, nausea: Double, Lumbar_pain:Double,Urine_pushing:Double,
Micturition_pains:Double,Burning_urethra:Double,decision:Double
)
//解析一行event内容,并映射为Diag类
def parseTravel(line: Array[Double]): Diag = {
Diag(
line(0), line(1) , line(2) , line(3) , line(4) , line(5) , line(6), line(7)
)
}
//RDD转换函数:解析一行文件内容从String,转变为Array[Double]类型,并过滤掉缺失数据的行
def parseRDD(rdd: RDD[String]): RDD[Array[Double]] = {
rdd.map(_.split(",")).map(_.map(_.toDouble)).filter(_.length==8)
}
def main(args: Array[String]) {
val conf = new SparkConf().setAppName("SparkDFebay").setMaster("local")
val sc = new SparkContext(conf)
val sqlContext = new SQLContext(sc)
import sqlContext.implicits._
//读文件并做简单转换
val data_path = "D:/code/flink-training-exercises-master/diagDataForRF/all.txt"
val diagDF = parseRDD(sc.textFile(data_path)).map(parseTravel).toDF().cache()
// diagDF.printSchema
//diagDF.show
//向量化处理---特征列向量化
val featureCols = Array("Temp", "what", "nausea","Lumbar_pain","Urine_pushing","Micturition_pains","Burning_urethra")
val assembler = new VectorAssembler().setInputCols(featureCols).setOutputCol("features")
val df2 = assembler.transform(diagDF)
//向量化处理---Label向量化
val labelIndexer = new StringIndexer().setInputCol("decision").setOutputCol("label")
val df3 = labelIndexer.fit(df2).transform(df2)
//数据集拆分为训练集,测试集
val splitSeed = 5043
val Array(trainingData, testData) = df3.randomSplit(Array(0.7, 0.3), splitSeed)
//定义算法分类器,并训练模型
val classifier = new RandomForestClassifier()
//定义评估器---多元分类评估(label列与Prediction列对比结果)
val evaluator = new MulticlassClassificationEvaluator().setLabelCol("label").setPredictionCol("prediction")
//定义算法变量调试范围,由于特征不是很多,子树设置范围不需要很大,这里设置成5-10
val paramGrid = new ParamGridBuilder()
.addGrid(classifier.maxBins, Array(25, 31))
.addGrid(classifier.maxDepth, Array(5, 10))
.addGrid(classifier.numTrees, Array(5, 10))
.addGrid(classifier.impurity, Array("entropy", "gini"))
.build()
//定义算法pipeline ,stage集合
val steps: Array[PipelineStage] = Array(classifier)
val pipeline = new Pipeline().setStages(steps)
//定义交叉验证器CrossValidator
val cv = new CrossValidator()
.setEstimator(pipeline)
.setEvaluator(evaluator)
.setEstimatorParamMaps(paramGrid)
.setNumFolds(10)
//pipeline执行,开始训练模型
val pipelineFittedModel = cv.fit(trainingData)
//对测试数据做预测
val predictions = pipelineFittedModel.transform(testData)
predictions.show(10)
//使用评估器评估模型精准度
val accuracy = evaluator.evaluate(predictions)
println("accuracy pipeline fitting:" + accuracy)
//println(pipelineFittedModel.bestModel.asInstanceOf[org.apache.spark.ml.PipelineModel].stages(0))
//模型保存
if (accuracy >0.8) {
try {
pipelineFittedModel.write.overwrite().save("./model/spark-LR-model-diag")
} catch {
case ex: Exception => println(ex)
case ex: Throwable => println("found a unknown exception" + ex)
}
}
}
}
向量化后的dataframe结构样例:
+----+----+------+-----------+-------------+-----------------+---------------+--------+--------------------+-----+--------------------+--------------------+----------+
|Temp|what|nausea|Lumbar_pain|Urine_pushing|Micturition_pains|Burning_urethra|decision| features|label| rawPrediction| probability|prediction|
+----+----+------+-----------+-------------+-----------------+---------------+--------+--------------------+-----+--------------------+--------------------+----------+
|35.0| 5.0| 0.0| 1.0| 0.0| 0.0| 0.0| 2.0|(7,[0,1,3],[35.0,...| 2.0| [0.0,0.0,8.0,0.0]| [0.0,0.0,1.0,0.0]| 2.0|
|36.0| 0.0| 0.0| 0.0| 1.0| 1.0| 1.0| 0.0|[36.0,0.0,0.0,0.0...| 0.0| [8.0,0.0,0.0,0.0]| [1.0,0.0,0.0,0.0]| 0.0|
|36.0| 2.0| 0.0| 0.0| 1.0| 1.0| 1.0| 0.0|[36.0,2.0,0.0,0.0...| 0.0| [8.0,0.0,0.0,0.0]| [1.0,0.0,0.0,0.0]| 0.0|
效果最好的RF 只需要5颗子树就可以了:
accuracy pipeline fitting:0.98
RandomForestClassificationModel (uid=rfc_cdec3565d92d) with 5 trees
模型训练–聚类:
def main(args: Array[String]): Unit = {
val conf = new SparkConf().setAppName("SparkDFebay").setMaster("local")
val sc = new SparkContext(conf)
val file="D:/code/flink/diagDataForRF/all.txt"
val rawTrainingData = sc.textFile(file)
val RDDData = rawTrainingData.map(line => {
Vectors.dense(line.split(",").map(_.trim).filter(!"".equals(_)).map(_.toDouble))
}).cache()
val Array(trainingData, testData) = RDDData.randomSplit(Array(0.7, 0.3))
//选取最合适的分类数
val ks: Array[Int] = Array(3, 4, 5, 6, 7, 8, 9, 10, 11, 12)
var ret:Map[Int,Double] =Map()
ks.foreach(cluster => {
val model: KMeansModel = KMeans.train(trainingData, cluster, 30, 1)
val ssd = model.computeCost(trainingData)
ret += (cluster->ssd)
})
ret.foreach{ i =>
println( "Key = " + i )}
}各聚类效果打印,[3-12]个聚类中,11个聚类的效果相对较好…
Key = (5,339.0946938424486)
Key = (10,200.3311688311819)
Key = (6,295.1888537187551)
Key = (9,218.09386154782624)
Key = (12,162.80075414781413)
Key = (7,277.8533613445493)
Key = (3,550.1058201058295)
Key = (11,160.2077922078047)
Key = (8,250.82946336430086)
Key = (4,378.9559529059061)
没有评论