项目说明:
心脏疾病预测
数据源
https://www.kaggle.com/sarubhai56/heart-disease
> 1. age > 2. sex > 3. chest pain type (4 values) > 4. resting blood pressure > 5. serum cholestoral in mg/dl > 6. fasting blood sugar > 120 mg/dl > 7. resting electrocardiographic results (values 0,1,2) > 8. maximum heart rate achieved > 9. exercise induced angina > 10. oldpeak = ST depression induced by exercise relative to rest > 11. the slope of the peak exercise ST segment > 12. number of major vessels (0-3) colored by flourosopy > 13. thal: 3 = normal; 6 = fixed defect; 7 = reversable defect 14. target: heart disease type
| age | sex | cp | trestbps | chol | fbs | restecg | thalach | exang | oldpeak | slope | ca | thal | target | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 63 | 1 | 3 | 145 | 233 | 1 | 0 | 150 | 0 | 2.3 | 0 | 0 | 1 | 1 |
| 1 | 37 | 1 | 2 | 130 | 250 | 0 | 1 | 187 | 0 | 3.5 | 0 | 0 | 2 | 1 |
| 2 | 41 | 0 | 1 | 130 | 204 | 0 | 0 | 172 | 0 | 1.4 | 2 | 0 | 2 | 1 |
数据处理:
数据分布情况—数值型:
里面的unique个数需要-1
age sex cp trestbps chol ... oldpeak slope ca thal target count 304 304 304 304 304 ... 304 304 304 304 304 unique 42 3 5 50 153 ... 41 4 6 5 3 top 58 1 0 120 204 ... 0 2 0 2 1 freq 19 207 143 37 6 ... 99 142 175 166 165
数据分布情况–非数值型:(不涉及)
各特征类型和非空情况统计:
基本上都是数值型数据
数据质量较好,因为是实时数据,后续还是做空值判断处理
Data columns (total 14 columns):
age 303 non-null int64
sex 303 non-null int64
cp 303 non-null int64
trestbps 303 non-null int64
chol 303 non-null int64
fbs 303 non-null int64
restecg 303 non-null int64
thalach 303 non-null int64
exang 303 non-null int64
oldpeak 303 non-null float64
slope 303 non-null int64
ca 303 non-null int64
thal 303 non-null int64
target 303 non-null int64
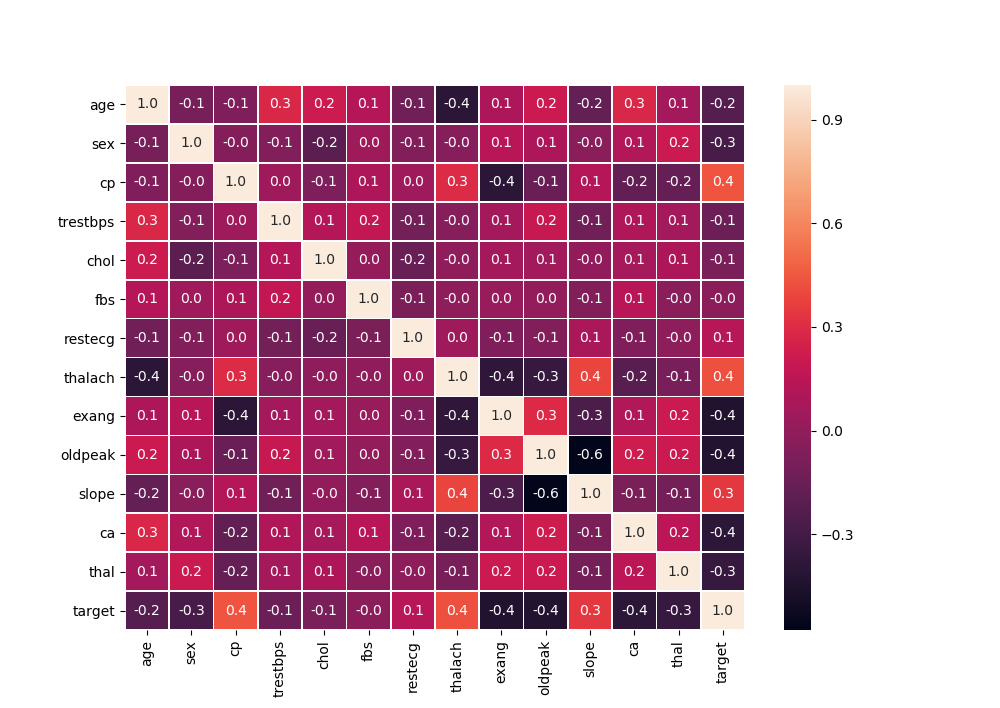
特征相关性—数值型
和target特别相关的特征不太多… 模型精准度可能会受影响
而且不适合在flink流程中使用simpleRegression方式做相关性空值实时预测,可以考虑使用均值做空值预测;
特征相关性—非数值型
不涉及
特征数据处理:
主要处理内容:
- 空值处理:空值比较少,可以考虑删除或者均值填充
- 异常值处理— 与均值差别太大的值删除
- 个别变量需要哑变量化处理:
age :分类成少年,青年,中年,老年几个阶段
chest pain type: 分成4列标签
electrocardiographic results: 按照值分为3类
the slope of the peak exercise ST segment:根据值的种类分成4列
thal:根据值的种类分成4列
主要通过使用flink 的state 特性,实现上述功能
数据结构类定义:
public class DataHeart implements Serializable {
public Integer age;
public Integer sex;
public Integer chestPain;
public Integer bloodPressure;
public Integer serumCholestoral;
public Integer bloodSugar;
public Integer electrocardiographic;
public Integer maximumHeartRate;
public Integer angina;
public Float oldpeak;
public Integer peakExerciseSTSegment;
public Integer numberOfMajorVessels;
public Integer thal;
public Integer target;
public DateTime eventTime;
public DataHeart() {
this.eventTime = new DateTime();
}
public DataHeart(int age, int sex, int chestPain, int bloodPressure, int serumCholestoral, int bloodSugar, int electrocardiographic,
int maximumHeartRate, int angina, Float oldpeak, int peakExerciseSTSegment, int numberOfMajorVessels, int thal, int target) {
this.eventTime = new DateTime();
this.age = age;
this.sex = sex;
this.chestPain = chestPain;
this.bloodPressure = bloodPressure;
this.serumCholestoral = serumCholestoral;
this.bloodSugar = bloodSugar;
this.electrocardiographic = electrocardiographic;
this.maximumHeartRate = maximumHeartRate;
this.angina = angina;
this.oldpeak = oldpeak;
this.peakExerciseSTSegment = peakExerciseSTSegment;
this.numberOfMajorVessels = numberOfMajorVessels;
this.thal = thal;
this.target = target;
}
public String toString() {
StringBuilder sb = new StringBuilder();
sb.append(age).append(",");
sb.append(sex).append(",");
sb.append(chestPain).append(",");
sb.append(bloodPressure).append(",");
sb.append(serumCholestoral).append(",");
sb.append(bloodSugar).append(",");
sb.append(electrocardiographic).append(",");
sb.append(maximumHeartRate).append(",");
sb.append(angina).append(",");
sb.append(oldpeak).append(",");
sb.append(peakExerciseSTSegment).append(",");
sb.append(numberOfMajorVessels).append(",");
sb.append(thal).append(",");
sb.append(target);
return sb.toString();
}
public static DataHeart instanceFromString(String line) {
String[] tokens = line.split(",");
if (tokens.length != 14) {
System.out.println("#############Invalid record: " + line+"\n");
//return null;
//throw new RuntimeException("Invalid record: " + line);
}
DataHeart diag = new DataHeart();
try {
diag.age = tokens[0].length() > 0 ? Integer.parseInt(tokens[0]):null;
diag.sex = tokens[1].length() > 0 ? Integer.parseInt(tokens[1]):null;
diag.chestPain = tokens[2].length() > 0 ? Integer.parseInt(tokens[2]) : null;
diag.bloodPressure = tokens[3].length() > 0 ? Integer.parseInt(tokens[3]) : null;
diag.serumCholestoral = tokens[4].length() > 0 ? Integer.parseInt(tokens[4]) : null;
diag.bloodSugar = tokens[5].length() > 0 ? Integer.parseInt(tokens[5]) : null;
diag.electrocardiographic =tokens[6].length() > 0 ? Integer.parseInt(tokens[6]) : null;
diag.maximumHeartRate = tokens[7].length() > 0 ? Integer.parseInt(tokens[7]) : null;
diag.angina = tokens[8].length() > 0 ? Integer.parseInt(tokens[8]) : null;
diag.oldpeak = tokens[9].length() > 0 ? Float.parseFloat(tokens[9]) : null;
diag.peakExerciseSTSegment = tokens[10].length() > 0 ? Integer.parseInt(tokens[10]) : null;
diag.numberOfMajorVessels = tokens[11].length() > 0 ? Integer.parseInt(tokens[11]) : null;
diag.thal = tokens[12].length() > 0 ? Integer.parseInt(tokens[12]) : null;
diag.target = tokens[13].length() > 0 ? Integer.parseInt(tokens[13]) : null;
} catch (NumberFormatException nfe) {
throw new RuntimeException("Invalid record: " + line, nfe);
}
return diag;
}
public long getEventTime() {
return this.eventTime.getMillis();
}
}
定义Source
由于大部分source内容类似,我们抽象出一个BaseSource类,大部分相同的操作都在该类中实现:
public class BaseSource<T> implements SourceFunction<T> {
protected final String dataFilePath;
protected final int servingSpeed;
protected transient BufferedReader reader;
protected transient InputStream FStream;
public BaseSource(String dataFilePath) {
this(dataFilePath, 1);
}
public BaseSource(String dataFilePath, int servingSpeedFactor) {
this.dataFilePath = dataFilePath;
this.servingSpeed = servingSpeedFactor;
}
public long getEventTime(DataHeart diag) {
return diag.getEventTime();
}
@Override
public void run(SourceContext<T> sourceContext) throws Exception {
}
@Override
public void cancel() {
try {
if (this.reader != null) {
this.reader.close();
}
if (this.FStream != null) {
this.FStream.close();
}
} catch (IOException ioe) {
throw new RuntimeException("Could not cancel SourceFunction", ioe);
} finally {
this.reader = null;
this.FStream = null;
}
}
}
定义具体的实现source类,重点run方法的重写:
public class HeartSource extends BaseSource<DataHeart> {
public HeartSource(String dataFilePath) {
super(dataFilePath, 1);
}
public HeartSource(String dataFilePath, int servingSpeedFactor) {
super(dataFilePath,servingSpeedFactor);
}
@Override
public void run(SourceContext<DataHeart> sourceContext) throws Exception {
super.FStream = new FileInputStream(super.dataFilePath);
super.reader = new BufferedReader(new InputStreamReader(super.FStream, "UTF-8"));
String line;
long time;
while (super.reader.ready() && (line = super.reader.readLine()) != null) {
DataHeart diag = DataHeart.instanceFromString(line);
if (diag == null){
continue;
}
time = getEventTime(diag);
sourceContext.collectWithTimestamp(diag, time);
sourceContext.emitWatermark(new Watermark(time - 1));
}
super.reader.close();
super.reader = null;
super.FStream.close();
super.FStream = null;
}
}
定义Operation Chain:
注意导入文件编码格式,一定要是utf-8格式编码!
// set up streaming execution environment
StreamExecutionEnvironment env = StreamExecutionEnvironment.getExecutionEnvironment();
// operate in Event-time
env.setStreamTimeCharacteristic(TimeCharacteristic.EventTime);
// start the data generator
DataStream<DataHeart> DsHeart = env.addSource(
new HeartSource(input, servingSpeedFactor));
DataStream<String> modDataStrForLR = DsHeart
.filter(new NoneFilter())
.map(new mapTime()).keyBy(0)
.flatMap(new NullFillMean())//均值填充空值
.flatMap(new heartFlatMapForLR());
modDataStrForLR.writeAsText("./HeartDataForLR");
env.execute("HeartData Prediction");
定义Map Operation类:
这里主要是为了后续使用AbstractRichFunction实现类需要,没有实际意义
public static class mapTime implements MapFunction<DataHeart, Tuple2<Long, DataHeart>> {
@Override
public Tuple2<Long, DataHeart> map(DataHeart energy) throws Exception {
long time = energy.eventTime.getMillis();;
return new Tuple2<>(time, energy);
}
}
定义部分特征空值过滤 Operation类:
public static class NoneFilter implements FilterFunction<DataHeart> {
@Override
public boolean filter(DataHeart heart) throws Exception {
return IsNotNone(heart.age) && IsNotNone(heart.angina)&& IsNotNone(heart.chestPain) ;
}
public boolean IsNotNone(Object Data){
if (Data == null )
return false;
else
return true;
}
}
定义空值预测Flatmap Operation 类:
对空值使用均值填充,这里使用ListState 结构保存多个特征的均值;
对bloodPressure,bloodSugar超出均值10 的数据删除,其他的用于继续累计均值;
public static class NullFillMean extends RichFlatMapFunction<Tuple2<Long, DataHeart> ,DataHeart> {
private transient ListState<Integer> heartMeanState;
private List<Integer> meansList;
@Override
public void flatMap(Tuple2<Long, DataHeart> val, Collector< DataHeart> out) throws Exception {
Iterator<Integer> modStateLst = heartMeanState.get().iterator();
Integer MeanBloodPressure=null;
Integer MeanBloodSugar=null;
if(!modStateLst.hasNext()){
MeanBloodPressure = 128;
MeanBloodSugar = 1;
}else{
MeanBloodPressure=modStateLst.next();
MeanBloodSugar=modStateLst.next();
}
meansList= new ArrayList<Integer>();
meansList.add(MeanBloodPressure);
meansList.add(MeanBloodSugar);
DataHeart heart = val.f1;
if(heart.bloodPressure == null){
heart.bloodPressure= MeanBloodPressure;
out.collect(heart);
}else if(heart.bloodSugar == null){
heart.bloodSugar= MeanBloodSugar;
out.collect(heart);
}else
{
if (abs(MeanBloodPressure-heart.bloodPressure)<10 && abs(MeanBloodSugar-heart.bloodSugar)<10) {
heartMeanState.update(meansList);
out.collect(heart);
}
}
}
@Override
public void open(Configuration config) {
ListStateDescriptor<Integer> descriptor2 =
new ListStateDescriptor<>(
// state name
"regressionModel",
// type information of state
TypeInformation.of(Integer.class));
heartMeanState = getRuntimeContext().getListState(descriptor2);
}
}定义 保存特定列特定文件格式的flatmap operation:
兼顾 部分数值特征 哑变量化功能
public static class heartFlatMapForLR implements FlatMapFunction<DataHeart, String> {
@Override
public void flatMap(DataHeart InputDiag, Collector<String> collector) throws Exception {
DataHeart heart = InputDiag;
DataHeartWashed heartWashed= new DataHeartWashed();
StringBuilder sb = new StringBuilder();
//sb.append(diag.date).append(",");
if (heart.age<20){
heartWashed.ageTeenage=1;
}else if(heart.age<30){
heartWashed.ageYoung=1;
}else if(heart.age<50){
heartWashed.ageMid=1;
}else {
heartWashed.ageOld=1;
}
if (heart.chestPain==0){
heartWashed.chestPain0=1;
}else if(heart.chestPain==1){
heartWashed.chestPain1=1;
}else if(heart.chestPain==2){
heartWashed.chestPain2=1;
}else {
heartWashed.chestPain3=1;
}
if (heart.electrocardiographic==0){
heartWashed.electrocardiographic0=1;
}else if(heart.electrocardiographic==1){
heartWashed.electrocardiographic1=1;
}else{
heartWashed.electrocardiographic2=1;
}
if (heart.peakExerciseSTSegment==0){
heartWashed.slope0=1;
}else if(heart.peakExerciseSTSegment==1){
heartWashed.slope1=1;
}else{
heartWashed.slope2=1;
}
if (heart.thal==0){
heartWashed.thal0=1;
}else if(heart.thal==1){
heartWashed.thal1=1;
}else if(heart.thal==1){
heartWashed.thal2=1;
}else{
heartWashed.thal3=1;
}
sb.append(heartWashed.ageTeenage).append(",");
sb.append(heartWashed.ageYoung).append(",");
sb.append(heartWashed.ageMid).append(",");
sb.append(heartWashed.ageOld).append(",");
sb.append(heart.sex).append(",");
sb.append(heartWashed.chestPain0).append(",");
sb.append(heartWashed.chestPain1).append(",");
sb.append(heartWashed.chestPain2).append(",");
sb.append(heartWashed.chestPain3).append(",");
sb.append(heart.bloodPressure).append(",");
sb.append(heart.serumCholestoral).append(",");
sb.append(heart.bloodSugar).append(",");
sb.append(heartWashed.electrocardiographic0).append(",");
sb.append(heartWashed.electrocardiographic1).append(",");
sb.append(heartWashed.electrocardiographic2).append(",");
sb.append(heart.maximumHeartRate).append(",");
sb.append(heart.angina).append(",");
sb.append(heart.oldpeak).append(",");
sb.append(heartWashed.slope0).append(",");
sb.append(heartWashed.slope1).append(",");
sb.append(heartWashed.slope2).append(",");
sb.append(heart.numberOfMajorVessels).append(",");
sb.append(heartWashed.thal0).append(",");
sb.append(heartWashed.thal1).append(",");
sb.append(heartWashed.thal2).append(",");
sb.append(heartWashed.thal3).append(",");
sb.append(heart.target);
collector.collect(sb.toString());
}
}
为了方便操作定义了一个简单的类:
只存储哑变量
public class DataHeartWashed implements Serializable {
public Integer ageTeenage;
public Integer ageYoung;
public Integer ageMid;
public Integer ageOld;
public Integer chestPain0;
public Integer chestPain1;
public Integer chestPain2;
public Integer chestPain3;
public Integer electrocardiographic0;
public Integer electrocardiographic1;
public Integer electrocardiographic2;
public Integer slope0;
public Integer slope1;
public Integer slope2;
public Integer thal0;
public Integer thal1;
public Integer thal2;
public Integer thal3;
public DataHeartWashed() {
this.ageTeenage = 0;
this.ageYoung = 0;
this.ageMid = 0;
this.ageOld = 0;
this.chestPain0 = 0;
this.chestPain1 = 0;
this.chestPain2 = 0;
this.chestPain3 = 0;
this.electrocardiographic0 = 0;
this.electrocardiographic1 = 0;
this.electrocardiographic2 = 0;
this.slope0 = 0;
this.slope1 = 0;
this.slope2 = 0;
this.thal0 = 0;
this.thal1 = 0;
this.thal2 = 0;
this.thal3 = 0;
}
}
保存到CSV文件中样例:
0,0,0,1,1,1,0,0,0,120,177,0,0,1,0,140,0,0.4,0,0,1,0,0,0,0,1,1 0,0,0,1,0,0,0,1,0,160,360,0,1,0,0,151,0,0.8,0,0,1,0,0,0,0,1,1 0,0,1,0,1,0,0,1,0,138,257,0,1,0,0,156,0,0.0,0,0,1,0,0,0,0,1,1 0,0,0,1,1,0,1,0,0,134,201,0,0,1,0,158,0,0.8,0,0,1,1,0,0,0,1,1 0,0,0,1,1,0,1,0,0,160,246,0,0,1,0,120,1,0.0,0,1,0,3,0,1,0,0,0
Spark 定时任务训练模型:
这里使用DecisionTree模型
object HeartDecisionTree {
case class Diag(
ageTeenage: Double, ageYoung: Double, ageMid: Double, ageOld: Double, sex: Double, chestPain0: Double,chestPain1: Double,chestPain2: Double,chestPain3: Double,
bloodPressure:Double,serumCholestoral:Double,bloodSugar:Double,ed0:Double,ed1:Double,ed2:Double,
maximumHeartRate:Double,angina:Double,oldpeak:Double,slope0:Double,slope1:Double,slope2:Double,
ca:Double,thal0:Double,thal1:Double,thal2:Double,thal3:Double,target:Double
)
//解析一行event内容,并映射为Diag类
def parseTravel(line: Array[Double]): Diag = {
Diag(
line(0), line(1) , line(2) , line(3) , line(4), line(5), line(6), line(7), line(8), line(9), line(10),
line(11), line(12), line(13),line(14), line(15), line(16), line(17),line(18), line(19), line(20),
line(21), line(22), line(23), line(24), line(25), line(26)
)
}
//RDD转换函数:解析一行文件内容从String,转变为Array[Double]类型,并过滤掉缺失数据的行
def parseRDD(rdd: RDD[String]): RDD[Array[Double]] = {
//rdd.foreach(a=> print(a+"\n"))
val a=rdd.map(_.split(","))
a.map(_.map(_.toDouble)).filter(_.length==27)
}
def main(args: Array[String]) {
val conf = new SparkConf().setAppName("sparkIris").setMaster("local")
val sc = new SparkContext(conf)
val sqlContext = new SQLContext(sc)
import sqlContext.implicits._
val data_path = "D:/code/flink/HeartDataForLR/*"
val FareDF = parseRDD(sc.textFile(data_path)).map(parseTravel).toDF().cache()
println(FareDF.count())
val featureCols = Array("ageTeenage","ageYoung", "ageMid", "ageOld","sex",
"chestPain0", "chestPain1","chestPain2","chestPain3","bloodPressure",
"serumCholestoral","bloodSugar","ed0","ed1","ed2","maximumHeartRate",
"angina","oldpeak","slope0","slope1","slope2","ca",
"thal0","thal1","thal2","thal3")
val assembler = new VectorAssembler().setInputCols(featureCols).setOutputCol("features")
val df2 = assembler.transform(FareDF)
df2.show
val Array(trainingData, testData) = df2.randomSplit(Array(0.7, 0.3))
//val classifier = new RandomForestClassifier()
val classifier = new DecisionTreeClassifier()
.setLabelCol("target")
.setFeaturesCol("features")
.setImpurity("gini")
.setMaxDepth(5)
val model = classifier.fit(trainingData)
try {
// model.write.overwrite().save("D:\\code\\spark\\model\\spark-LF-Occupy")
// val sameModel = DecisionTreeClassificationModel.load("D:\\code\\spark\\model\\spark-LF-Occupy")
val predictions = model.transform(testData)
predictions.show()
val evaluator = new BinaryClassificationEvaluator().setLabelCol("target").setRawPredictionCol("prediction")
val accuracy = evaluator.evaluate(predictions)
println("accuracy fitting: " + accuracy)
}catch{
case ex: Exception =>println(ex)
case ex: Throwable =>println("found a unknown exception"+ ex)
}
}
}
特征化后的df打印:
+----------+--------+------+------+---+----------+----------+----------+----------+-------------+----------------+----------+---+---+---+----------------+------+-------+------+------+------+---+-----+-----+-----+-----+------+--------------------+-------------+-----------+----------+
|ageTeenage|ageYoung|ageMid|ageOld|sex|chestPain0|chestPain1|chestPain2|chestPain3|bloodPressure|serumCholestoral|bloodSugar|ed0|ed1|ed2|maximumHeartRate|angina|oldpeak|slope0|slope1|slope2| ca|thal0|thal1|thal2|thal3|target| features|rawPrediction|probability|prediction|
+----------+--------+------+------+---+----------+----------+----------+----------+-------------+----------------+----------+---+---+---+----------------+------+-------+------+------+------+---+-----+-----+-----+-----+------+--------------------+-------------+-----------+----------+
| 0.0| 0.0| 0.0| 1.0|0.0| 0.0| 0.0| 1.0| 0.0| 140.0| 308.0| 0.0|1.0|0.0|0.0| 142.0| 0.0| 1.5| 0.0| 0.0| 1.0|1.0| 0.0| 0.0| 0.0| 1.0| 1.0|(26,[3,7,9,10,12,...| [0.0,16.0]| [0.0,1.0]| 1.0|
| 0.0| 0.0| 0.0| 1.0|0.0| 1.0| 0.0| 0.0| 0.0| 150.0| 244.0| 0.0|0.0|1.0|0.0| 154.0| 1.0| 1.4| 0.0| 1.0| 0.0|0.0| 0.0| 0.0| 0.0| 1.0| 0.0|(26,[3,5,9,10,13,...| [4.0,0.0]| [1.0,0.0]| 0.0|
模型效果评估打印:
accuracy fitting: 0.8263888888888888
没有评论